Phoenixin-14 alters transcriptome and steroid profiles in female green-spotted puffer (Dichotomyctere nigroviridis) | Scientific Reports - Nature.com
Abstract
Phoenixin (PNX) is a highly conserved, novel hormone with diverse functions, including hypothalamic control of reproduction, appetite modulation, and regulation of energy metabolism and inflammation. While some functions appear conserved across vertebrates, additional research is required to fully characterize these complex pleiotropic effects. For instance, very little is known about transcriptome level changes associated with PNX exposure, including responses in the hypothalamic–pituitary–gonadal (HPG) axis, which is critical in vertebrate reproduction. In addition, the PNX system may be especially complex in fish, where an additional receptor is likely present in some species. The purpose of this study was to assess hypothalamic and ovarian transcriptomes after PNX-14 administration in female vitellogenic green-spotted puffer (Dichotomyctere nigroviridis). Steroid-related changes were also assessed in the liver and blood plasma. Hypothalamic responses included pro-inflammatory signals such as interleukin 1β, possibly related to gut–brain axis functions, as well as suppression of cell proliferation. Ovarian responses were more widely downregulated across all identified pathways, which may reflect progression to a less transcriptionally active state in oocytes. Both organs shared regulation in transforming growth factor-β and extracellular matrix remodeling (periostin) pathways. Reproductive processes were in general downregulated, but both inhibiting (bone morphogenetic protein 15 and follistatin) and promoting (17-hydroxyprogesterone) factors for oocyte maturation were identified. Select genes involved in reproduction (vitellogenins, estrogen receptors) in the liver were unresponsive to PNX-14 and higher doses may be needed to induce reproductive effects in D. nigroviridis. These results reinforce the complexity of PNX actions in diverse tissues and highlight important roles for this hormone in regulating the immune response, energy metabolism, and cell growth.
Introduction
Phoenixin (PNX) is a highly conserved, novel hormone first discovered using a bioinformatics approach to search the human genome for previously unknown secreted peptides1. PNX is a proteolytic cleavage product of small integral membrane protein 20 (SMIM20), also known as MITRAC7, which functions in respiratory chain assembly in mitochondria to chaperone cytochrome c oxidase2,3,4. The C-terminal domain of SMIM20 can be cleaved into several amidated forms, with the 14- and 20-amino acid peptides (PNX-14 and PNX-20, respectively) being the most predominant4. Both forms appear functionally similar and exhibit highest expression in the mammalian hypothalamus, with lower levels in the heart, thymus, and digestive system1,5. A putative receptor for PNX ligands was recently identified as GPR173, also known as SREB3, which is a member of the highly conserved G protein-coupled receptor family SREB (Super-conserved Receptors Expressed in Brain)6,7,8. PNX effects appear to require functional SREB3, although its status as the receptor is unclear since direct binding assays have been largely unsuccessful, and evolutionary studies may not support this relationship7,9. Overall, the PNX/SREB3 system has been associated with pleiotropic functions in numerous tissues, ranging from control of reproduction in the ovary and hypothalamus to stress and anxiety modulation, anti-inflammatory roles, appetite and thirst regulation, and glucose homeostasis4,10.
Some of the more well-studied functions of PNX are related to metabolism within the gut–brain axis. For example, intracerebroventricular PNX injection is associated with increased food intake in mice and rats and can lower core body temperature with anxiolytic effects11,12,13. Within stomach endocrine cells, PNX may stimulate ghrelin production and inhibit cholecystokinin, while pancreatic islet functions include insulin secretion and beta cell proliferation in rats14,15. Metabolic effects also extend beyond these sites, where PNX induces osteoblast differentiation in a mouse bone cell line and has anti-inflammatory properties in the small intestine16,17. Indeed, immune-related and proliferative roles may be critical in PNX/SREB3 signaling, since PNX promotes mitochondrial biogenesis and exhibits protective effects in the brain and heart18,19,20.
Despite recent research into PNX metabolic and immune functions, the hormone is likely most well known as a hypothalamic neuropeptide that promotes reproductive processes1. These functions include effects on the estrous cycle in rats, stimulation of gonadotropin releasing hormone (GnRH) and kisspeptin neurons in the hypothalamus, and luteinizing hormone (LH) release from the pituitary1,7,8. Overall, PNX seems broadly associated across the hypothalamic–pituitary–gonadal (HPG) axis and is also localized within the human ovary, inducing granulosa cell proliferation, estrogen production, and ovulation21. Some reproductive dysfunctions are also associated with the PNX/SREB3 system, including endometriosis, polycystic ovary syndrome in humans and rats, and cystic endometrial hyperplasia in dogs22,23,24,25.
The PNX/SREB3 system is far less-studied in non-mammalian vertebrates, but recent research has identified conserved roles in both appetite regulation and reproduction. For instance, hypothalamic PNX also affects food intake in chicken (Gallus gallus), spotted scat (Scatophagus argus), and zebrafish (Danio rerio)26,27,28. Many reproductive processes are also promoted by PNX in fish, including numerous gene expression increases in the hypothalamus, pituitary, ovary, and liver, as well as oocyte maturation in zebrafish29,30. However, data remains sparse for the role of PNX in vertebrates, especially in fish, where recent research identified a duplicated SREB3 gene (sreb3b) that is only found in some species31. The presence of a second SREB3 in PNX signaling may indicate the importance of this system in fish. Furthermore, to our knowledge, no study to date has assessed transcriptomic responses following PNX treatment in any vertebrate, which was recently identified as a research gap in our collective understanding of PNX signaling10. To this end, we determined the molecular effects of PNX-14 on the HPG axis in female green-spotted puffer (Dichotomyctere nigroviridis) using RNA-sequencing. PNX-14 was previously associated with reproductive changes in another perciform species29, and a dose was chosen (100 ng/g body weight) that is known to affect reproduction in two other fishes29,30. Puffer was chosen because it expresses both sreb3 genes31 and is characterized by reproductive dysfunction in aquaculture conditions, where ovarian development progresses to vitellogenesis, but does not undergo oocyte maturation without artificial hormonal intervention32. Given its conserved role in the HPG axis, PNX may therefore be a useful hormone to regulate oocyte maturation in diverse fishes. We assessed hypothalamic and ovarian transcriptomes, along with steroid-related analyses in blood plasma and liver, to: (1) broadly characterize PNX-14 transcriptome effects, and (2) determine if treatment affected reproductive processes associated with oocyte maturation in vitellogenic pufferfish.
Methods
PNX-14 treatments and puffer sampling
Sexually mature female green-spotted puffers (8.6 ± 0.3 g, 60.3 ± 0.65 mm total length) of wild origin were imported through a local wholesaler (Segrest Farms, Inc.) and held in a freshwater flow through system at the University of Florida Tropical Aquaculture Laboratory (Ruskin, FL, USA). All puffers were maintained and sampled under approved guidelines of the University of Florida (UF) Institutional Animal Care and Use Committee (IACUC #: 202011293). All experimental protocols were performed in accordance with the University of Florida IACUC ethical guidelines and regulations for vertebrate animal use, as well as approval through the University of Maine at Farmington (UMF) Grant Coordination Committee. All experiments were conducted in compliance with ARRIVE guidelines.
To promote ovarian development to the vitellogenic stage, fish were slowly acclimated to saltwater (30 g/L salinity) over 3 months32. Fish (n = 12) were then anesthetized with 150 mg/L neutral-buffered tricaine methanesulfonate (MS-222), confirmed to be vitellogenic by ovarian biopsy33, and intramuscularly injected with puffer-specific PNX-14 (100 ng/g body weight, n = 6) or sterile water control (n = 6). PNX was diluted using sterile water, and the 100 ng/g body weight concentration was previously used in both spotted scat and zebrafish with significant HPG axis effects29,30. Briefly, to identify puffer-specific PNX, a spotted scat (Scatophagus argus) sequence (GenBank Acc. No. MH360732)29 was compared using BLAST to the puffer genome on Ensembl. The significant match (GSTENT10013530001) was confirmed manually to be identical and highly similar to D. nigroviridis and Takifugu rubripes sequences, respectively, in NCBI databases. The sequence was translated using ORFfinder (NCBI), and the PNX-14 peptide (DVQPVGMKIWSDPF-amide) was purchased and met all quality standards (≥ 98% purity) from Genscript USA, Inc. (Piscataway, NJ, USA).
Following injection, PNX-14 treated and control fish were held in individual 20 L containers housed within two 400 L culture tanks within a 1400 L recirculating system for 24 h, then euthanized with 200 mg/L neutral-buffered MS-222. All 20 L containers had small openings to allow for passive water flow and homogenous water quality parameters among replicates. To assess reproductive changes in response to PNX-14, ovarian biopsies and hypothalamic brain sections34 were preserved in RNALater (Ambion, Inc., Austin, TX, USA), snap frozen on dry ice, and stored at − 80 °C until subsequent RNA extractions at UF. To assess steroid-related patterns in response to PNX-14, the experiment was repeated using a second set of fish (n = 6 fish/treatment) and all individuals were euthanized 24 h following PNX-14 exposure. Liver samples were preserved in RNALater (Ambion, Inc., Austin, TX, USA), snap frozen on dry ice, and stored at − 80 °C until shipment to UMF for subsequent RNA extractions. Blood samples were collected in lithium heparinized vials following caudal ablation. Samples were centrifuged, and separated blood plasma (3–22 μl/fish) was frozen (− 20 °C) prior to steroid quantification at the UF Analytical Toxicology Core Laboratory.
RNA preparation and RNA sequencing (RNA-seq)
Total RNA for each ovary and hypothalamus was isolated using TRIzol (Thermo Fisher Scientific, Waltham, MA USA) as per the manufacturer's instruction for transcriptomics. Due to budget constraints, we did not sequence the liver transcriptome but turned to a focused qPCR approach for transcripts important for reproduction. RNA concentration was determined using the Qubit® 2.0 Fluorometer (ThermoFisher/Invitrogen, Grand Island, NY, USA) and RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). A total of 22 RNA samples were deemed high quality for RNA-seq library construction and consisted of six control hypothalami (HC group), six treatment hypothalami (HT), five control ovaries (OC), and five treatment ovaries (OT). Total RNA was used for mRNA isolation using the NEBNext Ploy(A) mRNA Magnetic Isolation module (New England Biolabs, catalog # E7490). RNA library construction was then performed with the NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina® (New England Biolabs, catalog #E7760) according to the manufacturer's user guide. Individually prepared libraries were pooled by equimolar concentrations and sequenced by NovoSeq 6000 using 150 bp paired end reads (Illumina Inc., CA, USA). Sequencing was performed by Novogene Corporation (Beijing, China). Raw data (raw reads) of FASTQ format were first processed through fastp. In this step, clean data (clean reads) were obtained from raw data by removing reads containing adapter and poly-N sequences and reads with low quality. At the same time, Q20, Q30 and GC content of the clean data were determined (Supplementary Table S1). All downstream analyses were based on clean data with high quality.
The reference genome (TETRAODON 8.0) and gene model annotation files were downloaded from the Ensembl genome website browser directly. Paired-end clean reads were mapped to the reference genome using HISAT2 software. Feature counts were used to count the read numbers mapped to each gene, including known and novel genes. Reads Per Kilobase of transcript, per Million mapped reads (RPKM) of each gene was calculated based on the length of the gene and read count mapped (Supplementary Fig. S1). Ovary and hypothalamus transcriptomes were submitted to the NCBI Gene Expression Omnibus (GEO) database (GSE183029). DESeq2 was used for differential expression analysis between the OC and OT and between HC and HT groups. Differentially expressed gene lists with log2 fold changes and P-values are provided in the Supplementary Data. To provide the most complete gene name information for downstream pathway analyses, each differentially expressed gene list was further annotated manually. Briefly, both ovary and hypothalamus genes initially listed as unknown and differentially expressed across treatments (P < 0.05) were manually used in BLAST searches against the nucleotide (nt) database (NCBI) to identify significant matches to known genes in other fishes (e-value < 1e−05). If ten or greater matches in the database to other fish species were present, then each unknown gene was provided with a gene ID using Zebrafish Information Network (ZFIN) nomenclature. The revised ovary and hypothalamus differentially expressed gene lists (Supplementary Data) were then used in pathway analyses.
Pathway analysis
Gene set enrichment analysis (GSEA) and subnetwork enrichment analysis (SNEA) were conducted in Pathway Studio 12.0 (Elsevier, Amsterdam, Netherlands). A total number of 12,766 ovary and 12,966 hypothalamus genes were successfully mapped to mammalian homologs using the official gene name. Gene set enrichment analysis proceeded with 1000 permutations to generate the distributions. Pathway Studio conducted statistical enrichment based upon ontologies and curated pathways. A two-sample nonparametric Kolmogorov–Smirnov test was used to compare the cumulative distributions of two data sets (networks) for differences. Subnetwork enrichment analysis or SNEA is designed around networks of common regulators of expression using known relationships (i.e., expression, binding, etc.) derived for experimental data and literature which are focused on gene hubs. A distribution of expression values is calculated by a permutation algorithm to obtain a "background" distribution followed by a statistical comparison between the sub-network data (query data) and the background distribution using a Mann–Whitney U test. For both GSEA and SNEA, a P-value is generated to indicate whether a process is statistically enriched in the query dataset relative to what is expected by random chance based upon the background distribution. Enrichment P-value for a gene seed was set at P < 0.05. GSEA and SNEA lists for both ovary and hypothalamus are provided in the Supplementary Data.
To broadly compare transcriptome responses across organs, gene sets common to both ovary and hypothalamus were identified, and highly significant sets were assessed by P < 0.01 and > 50% of total pathway entries measured in the dataset as significantly different. Highly significant sets were then manually assigned to broad functional categories, mostly based on information in the UniProt database (Ensembl, Hinxton, Cambridgeshire, UK). To further assess if organ transcriptional responses were associated with broad up- or downregulation, median fold changes of these gene sets were used in Chi-square tests with expected equal frequencies between the number of positive and negative median fold changes. To more fully characterize changes specific to reproductive processes, each organ dataset was also queried for significant gene differences associated with selected anatomical concepts or cell processes (e.g. ovarian follicle or reproduction, respectively). Lastly, for SNEA, highly significant subnetworks in both organs were characterized by > 60% of total neighbors measured in the dataset with P-value < 1e−05.
Liver RNA extractions and cDNA synthesis
Liver RNA extractions were performed at UMF using Tri Reagent (Sigma-Aldrich, St. Louis, MO, USA) and standard phenol/chloroform procedures, followed by a third precipitation step using polyvinylpyrolidone (PVP) (2% PVP, 1.4 M NaCl) and 5 M LiCl to remove polysaccharide contamination35. Total RNA quantity and quality were assessed using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and 1.0% agarose gel electrophoresis. Total RNA (2.5 μg/sample) was treated with ezDNase and a 5 min incubation at 37 °C to remove genomic DNA contamination (Invitrogen, Carlsbad, CA, USA). In addition, ovary and hypothalamus total RNA previously extracted at UF were treated with TURBO™ DNase following manufacturer's protocols (Thermo Fisher Scientific) and shipped frozen to UMF for later quantitative PCR (qPCR) confirmation of RNA-seq differential gene expression. Ovary and hypothalamus sample sizes were n = 3, 4, 5, and 6 for OC, OT, HC, and HT groups, respectively. Liver, ovary, and hypothalamus cDNA synthesis was conducted using the Superscript IV VILO kit (Invitrogen).
Quantitative PCR (qPCR)
To confirm RNA-seq identified expression patterns, ovary and hypothalamus cDNAs were used in qPCR assays with a StepOne Plus Real Time PCR System and FAST SYBR™ Green Master Mix (Applied Biosystems, Waltham, MA, USA). Primers were designed using NCBI Primer-BLAST from genes of interest identified in the transcriptomes (Supplementary Table S2). A total of three hypothalamus genes (dkk3a, dpydb, and tgfb3) and seven ovary genes (acta1, dpydb, isr2a, safb, slc31a1, slc26a2, and srl) were assessed in qPCR. To assess steroid-related expression changes in the liver, six genes were assayed including estrogen receptors (esr1, esr2a, esr2b), sex hormone binding globulin (shbg), and vitellogenin genes (vtgab and vtgc), based on previous research in zebrafish30. Primers for liver genes (Supplementary Table S2) were designed similarly to above from sequences available in the D. nigroviridis genome (Ensembl) (ENSTNIT00000015435, ENSTNIT00000005784, ENSTNIT00000015555, ENSTNIT00000008218, ENSTNIT00000000661, and ENSTNIT00000003224, respectively). Primer sets were verified for intended product amplification using standard PCR and 2.0% agarose gel electrophoresis, following prior protocols36. Assays were normalized using eef1a, which did not exhibit significant differences in expression across treatments (P > 0.05). All qPCR assays consisted of 10 μl reaction volumes, 1.33 μl diluted template, 0.02–1.0 μM primers (depending on assay), and standard cycling conditions (95 °C for 10 min, 40 cycles of 95 °C for 15 s and 60 °C for 1 min). All samples were assayed in duplicate, and relative standard curves (run in triplicate) were made from pooled cDNA. Optimized linear standard curves consisted of four to six points, with approximately 90–100% PCR efficiency and single peak amplification in dissociation curve analysis. Standard qPCR negative controls (no template and no reverse transcriptase) were also used and exhibited no contamination (liver) or only negligible levels (most Ct > 30, and five or more cycles greater than most samples) in ovary/hypothalamus assays.
For all assays, qPCR results were expressed relative to the control (set to 1.0) and analyzed using relative quantification37. Data were expressed as mean ± standard error, and log-transformed relative expression levels were analyzed using two sample t tests in SYSTAT12 (Systat Software Inc., San Jose, CA, USA) and assessed at the P < 0.05 level. To identify correlations (P < 0.05) between RNA-seq normalized count data and relative qPCR expression, least squares linear regression analyses (SYSTAT12) were also conducted38.
Plasma steroid quantifications
Blood plasma steroid levels were quantified using a liquid chromatography tandem mass spectrometry (LC–MS/MS) method at the UF Analytical Toxicology Core Laboratory previously validated for low volumes39. Briefly, a QTRAP 6500 linear ion quadrupole LC–MS/MS mass spectrometer (AB Sciex, Framingham, MA, USA) was used with a Shimadzu Scientific Instruments (Columbia, MD, USA) liquid chromatography (LC) system and a Kinetex© PS C18 column (2.6 μm particle size, 100 Å pore size, and 2.1 × 150 mm diameters). Injection volumes were 10 μl. Six steroids were quantified (progesterone, 17-hydroxyprogesterone, corticosterone, cortisol, testosterone, and 11-ketotestosterone). Estradiol (E2) was not measured due to low overall concentrations. Steroid concentrations (ng/μl plasma) were log-transformed, assessed for normality, and used in two-sample t tests in SYSTAT12 to detect significant differences (P < 0.05) between treatments.
Results
Ovary and hypothalamus transcriptomes
Ovary and hypothalamus transcriptomes were overall high quality, generating 20.2–27.0 million raw reads per sample that were processed to 19.8–26.5 million clean reads per sample (Supplementary Table S1). Error rates were low for all samples (0.03%) and Q20 and Q30 quality scores were high (mean 97.5% and 93.3%, respectively). Mapping rates to the puffer reference genome were also high (mean 90.0% and 85.5% total and uniquely mapping rates, respectively) and fragments per kilobase of transcript per million mapped reads (FPKM) levels were similar across samples (Supplementary Fig. S1). Mapping identified 20,562 total unique gene sequences across both organ types. Ovary and hypothalamus transcriptome profiles exhibited high divergence from each other but overall low discrimination between control and PNX-treated samples within an organ type (Fig. 1A). In addition, hypothalamus samples were separated into two groups that did not reflect treatments (HC and HT labels), although the variation along this component was far less (2.32%) than that between organs (86.92%). Differential gene expression analysis was used to identify 36 total genes with highly significant (P adj. < 0.05) up- or downregulation following PNX-14 treatment, which were divided into 31 ovary genes and five hypothalamus genes (Fig. 1B, see Supplementary Data for gene lists). Ovary samples exhibited clearer discrimination by treatment among highly significant genes (Fig. 1B, top blue and orange labels), while hypothalamus samples overall did not group by treatment. Among all differentially expressed genes (raw P-value < 0.05), the ovary and hypothalamus exhibited totals of 1206 and 582 genes, respectively, with 113 common genes between organ types (Fig. 1C). Following pathway analyses, these gene sets were categorized into 216 and 250 unique pathways with significant up- or downregulation (P < 0.05) in the ovary and hypothalamus, respectively (Fig. 1D). A total of 78 significantly different pathways were shared between organ types (Supplementary Data).
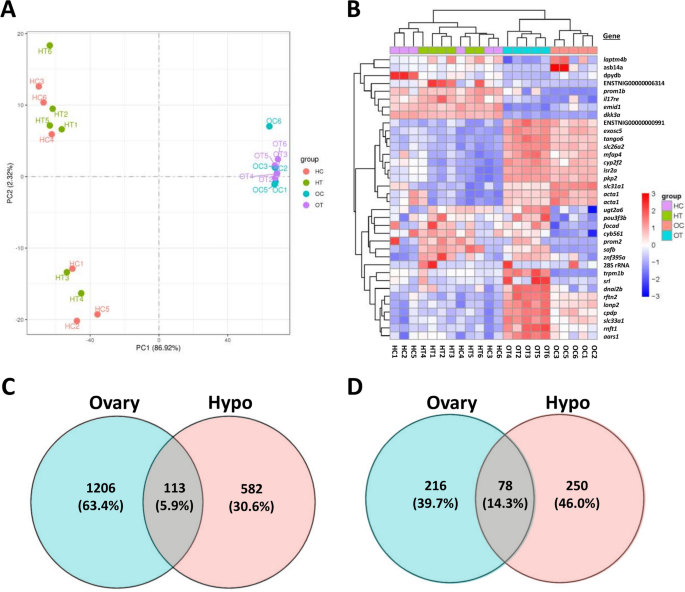
Principal component analysis (A) of transcriptomes in ovary control (OC, blue), ovary PNX-14 treated (OT, purple), hypothalamus control (HC, red) and hypothalamus PNX-14 treated (HT, green) samples. (B) Heat map of 36 differentially expressed genes (false discovery rate < 0.05) across both tissues, organized by genes (rows) and samples (columns). Gene names and individual sample IDs are provided on the right and bottom, respectively. Red colors indicate positive log2 fold changes in expression, while blue indicates negative changes. Unknown genes are labeled with their respective gene IDs identified in the Ensembl puffer genome (TETRAODON 8.0). (C) Total differentially expressed genes (P < 0.05) or (D) pathways with percentages (%) in the PNX-14 treatment in ovary (blue) and hypothalamus (red) transcriptomes, with shared entries across both transcriptomes identified in the middle (purple).
Selected ovary and hypothalamus gene patterns were largely confirmed in qPCR (6 of 10 total assays, Fig. 2). Ovary patterns were variable, where three genes (srl, isr2a, and acta1) exhibited significant positive correlations between RNA-seq and qPCR data but did not have significant changes in relative expression (Fig. 2A). Four other ovary genes (safb, slc31a1, slc26a2, and dpydb) did not exhibit positive correlations (Supplementary Fig. S2), which were likely due to overall low sample sizes and fold changes for most genes (< 2.0). However, all three hypothalamus genes assayed (dkk3a, dpydb, and tgfb3) were confirmed across RNA-seq and qPCR techniques, with significant expression changes and positive correlations (Fig. 2B).
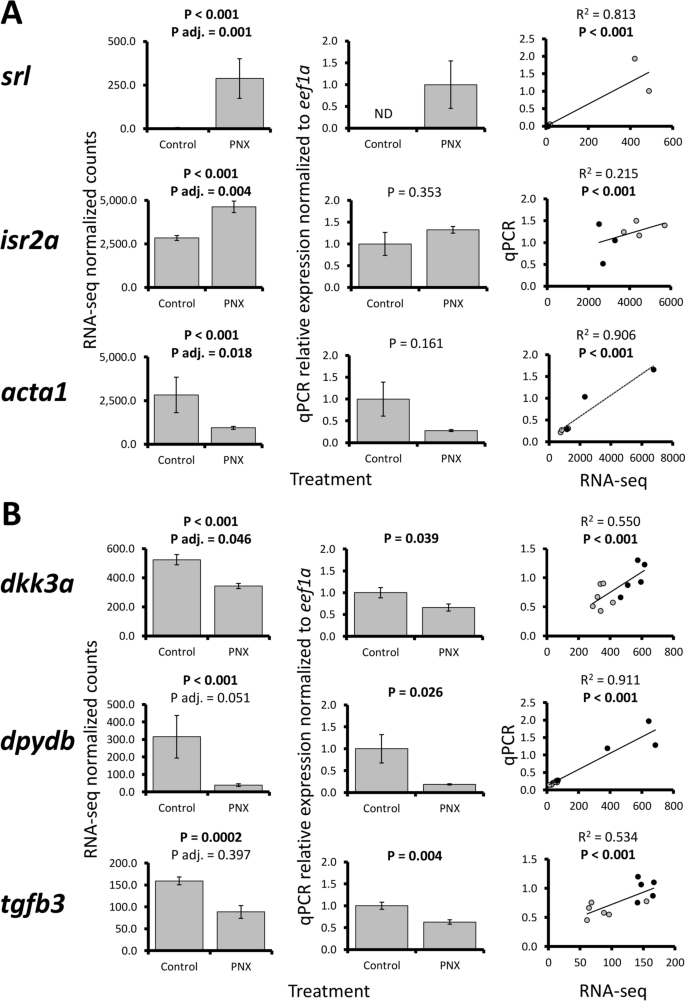
RNA-seq normalized count data (first column), relative mRNA expression in qPCR normalized to eef1a (second column), and linear regression analyses (third column) of selected ovary [(A) srl, isr2a, and acta1] and hypothalamus genes [(B) dkk3a, dpydb, and tgfb3]. Each bar represents the mean ± standard error and significance was assessed at P < 0.05. P adjusted (adj.) refers to the multiple test correction in RNA-seq differential expression analyses (false discovery rate < 0.05). For regression analyses, black and gray circles refer to control and PNX-14 treated samples, respectively.
Transcriptome networks
Both ovary and hypothalamus exhibited changes in numerous gene pathways following PNX exposure, including highly significant shifts in 54 and 74 gene sets, respectively (Supplementary Data). When these were grouped into functional themes, both organs exhibited a majority of changes likely related to immune responses, including natural killer cell activation, T cell receptor pathways, and cytokine signaling. Overall, 35 and 39 highly significant gene sets were associated with immune responses in the ovary and hypothalamus, respectively (Fig. 3). The second and third most abundant categories were also similar in theme across organs, with nine and 11 sets associated with growth factors (e.g. transforming growth factor-α and amphiregulin pathways), and four and six sets being cancer-related (e.g. "ERBB/VEGFR/Akt signaling in breast cancer" and "Hodgkin lymphoma"). Less abundant categories varied more between organs, but some were still shared such as insulin signaling, steroid signaling, apoptosis, and cell adhesion, even though the specific gene sets within those themes often varied. Other categories related to leptin signaling and thyroid function were unique to the ovary, while ion movement, chromosome condensation, diabetes-related, endothelin signaling, epigenetic modification, glucose metabolism, kinetochore assembly, mucin production, and retrotransposon-related were unique to the hypothalamus. Overall, even though some categories were shared between organs, the PNX response differed greatly. The ovary was characterized by entirely negative median fold changes in highly significant gene sets (P < 0.0001), while the hypothalamus exhibited mostly increases (P < 0.01), especially in immune response (Fig. 3).
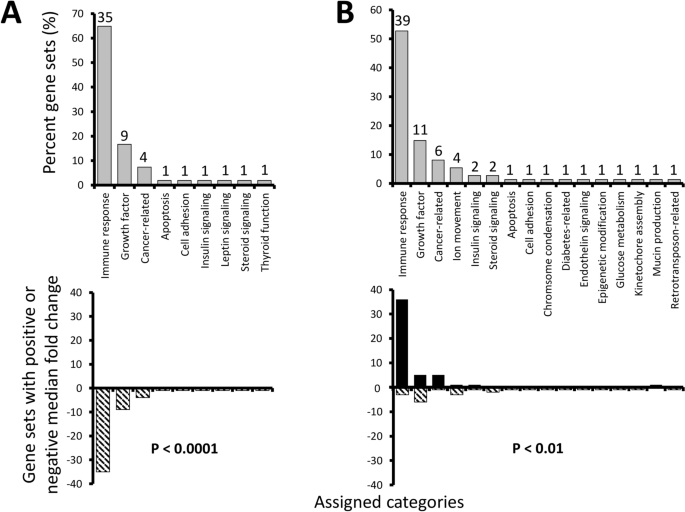
Percent gene sets (%) in the ovary (A) and hypothalamus (B) identified as highly significant (P < 0.01 with > 50% measured entries in a pathway) following PNX treatment were manually assigned functional categories (upper graphs). Numbers above gray bars refer to the total gene set number assigned to each category. Lower graphs identify the percent of gene sets (%) with overall positive (black bars) or negative (striped) median fold changes in each category. P values refer to significant differences from an expected equal frequency of gene sets with positive and negative changes.
Many highly significant gene sets were associated with known PNX roles in energy metabolism, nociception, reproduction, and inflammatory response (Table 1). These sets included "LIF Expression Targets" (− 1.16 median fold change) and "InsulinR → CTNNB/FOXA/FOXO Signaling" (− 1.14) in the ovary, and "IL1B Expression Targets → Nociception" (1.07) and "Estrogens in Prolactin Production" (− 1.05) in the hypothalamus. In particular, the "Chromosome Condensation" gene set was among the greatest hypothalamic median fold changes detected (− 1.22) and also exhibited 100% representation, with 21 genes exhibiting changes out of 21 total genes in the pathway (Table 1). Overall, most genes in the chromosome condensation network exhibited expression decreases, except for increases in serine-threonine kinase 4 (stk4) and non-SMC condensin I complex subunit G (ncapg) (Fig. 4).
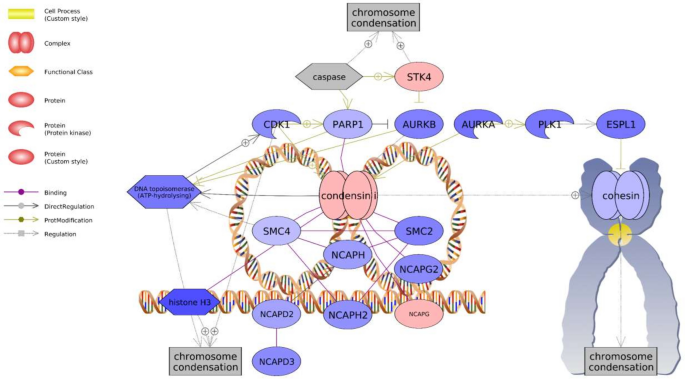
Chromosome condensation-associated network identified in the hypothalamus following PNX treatment. Red indicates an upregulated gene in the network (darker shade indicates greater log2 fold change compared to control), while blue indicates a downregulated gene. Shapes and arrows are identified in the legend to the left. This is a proposed mechanism based only on transcriptomic data.
Of all highly significant gene sets, only six were shared in both organs (Table 1). These included four likely immune response pathways ("gamma Globulins Expression Targets," "IL1B Expression Targets," "Natural Killer Cell Activation through ITAM-Containing Receptors," and "TCR → STAT Expression Targets") one growth factor-associated pathway ("TGFB1-TGFBR1/AP-1 Expression Targets"), and one cell adhesion pathway ("Periostin (POSTN) Production by Airway Epithelium in Asthma"). Five of these sets exhibited differences in median fold changes between organs, with the immune- and growth-related pathways more greatly decreasing in the ovary and slightly increasing in the hypothalamus. For example, the transforming growth factor-β (tgfb) associated network was characterized by more extensive downregulation in the ovary, with upstream decreases in both the tgfb1 ligand and a receptor (tgfbr2) (Supplementary Fig. S3). In contrast, the same network in the hypothalamus was characterized by increases in these genes, along with broad increases in many downstream targets (Supplementary Fig. S4). Only the postn-associated pathway exhibited similar changes across both organs (− 1.44 vs. − 1.07 downregulation) and was characterized by 76.5% of the total genes changing in response to treatment. The changes in this network were associated with decreases in tyrosine-protein kinases (tyk2, jak1, and jak3), a gene with similarity to proto-oncogene c-Fos (fos), periostin (postn), and several downstream targets (Fig. 5).
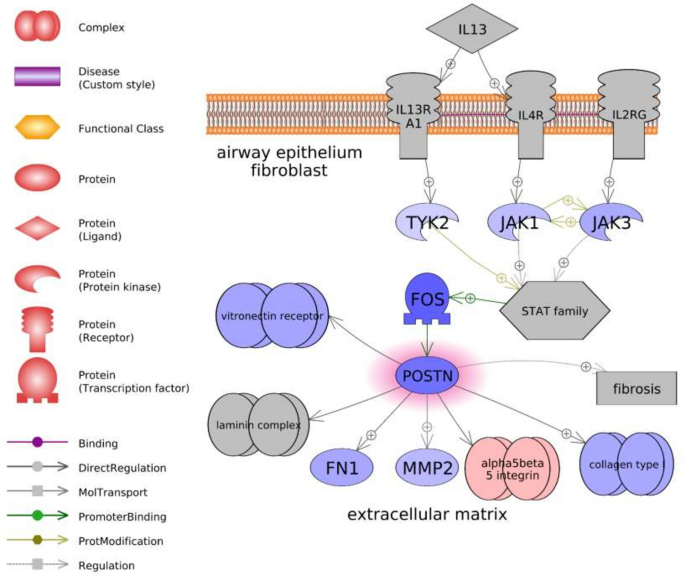
Periostin (postn)-associated network identified in the ovary following PNX treatment. Red indicates an upregulated gene in the network (darker shade indicates greater log2 fold change compared to control), while blue indicates a downregulated gene. Shapes and arrows are identified in the legend to the left. This is a proposed mechanism based only on transcriptomic data.
Both the ovary and hypothalamus also exhibited evidence of downregulation in reproductive processes. When the ovary transcriptomes were queried for genes associated with anatomical concepts in reproduction, a network was identified with 85 genes that were largely downregulated in response to PNX-14 (Supplementary Fig. S5). Of note, this network included increases in hormones associated with reproductive processes in the ovary (e.g. bone morphogenetic protein 15, bmp15; gonadotropin-releasing hormone 2, gnrh2) but decreases in many transcription factor genes. There were no changes in steroidogenic enzymes, except for an increase in 17-beta-hydroxysteroid dehydrogenase 7 (hsd17b7). In contrast, a smaller reproductive network (17 genes) was identified in the hypothalamus and was based on cell processes of reproduction, lactation, and apoptosis (Supplementary Fig. S6). This network was also largely downregulated and included decreases in a dopamine receptor (drd2), estrogen receptors (esr1 and esr2), prolactin (prl), and luteinizing hormone. Some growth factors associated with reproductive processes varied, such as increases in tgfb1 but decreases in transforming growth factor-β 3 (tgbf3) and fibroblast growth factor 2 (fgf2).
Regarding subnetworks, a total of 50 were both highly significant and shared across ovary and hypothalamus (see SNEA in Supplementary Data), with a smaller set associated with reproduction. These subnetworks were also downregulated and included regulators of hormone secretion (− 1.09 and − 1.02 median change in ovary and hypothalamus, respectively), fertilization (− 1.05 and − 1.02), and ovary follicle maturation (− 1.15 and − 1.00). Many other subnetworks were also shared between organs, with some of the most highly significant in both being regulators of chemotaxis (− 1.09 and 1.02) and cell interaction (− 1.10 and − 1.00). In addition, two subnetworks related to regulators of pregnancy (− 1.07 and − 1.01) and calcium mobilization (− 1.08 and 1.00) also exhibited high P-values in both organs (> 1e−08) but were less represented among measured neighbors (57–59%). Overall, other top subnetworks ranked by P-value in the ovary focused on decreases in neural signals (e.g. excitability, transmission of nerve impulse, and perception of pain) while top subnetworks in the hypothalamus were increases related to immune response (e.g. leukocyte activation, NK cell mediated cytotoxicity, and immune system function).
Steroid-related patterns in liver and blood plasma
PNX-14 treatment was overall associated with small changes in steroid-related patterns. In the liver, no significant expression changes were detected in vitellogenin genes or estrogen receptors (Fig. 6). However, a marginal but non-significant increase (< 2-fold) in shbg was detected following treatment (P = 0.067). Blood plasma steroid concentrations were overall low for progesterone, corticosterone, cortisol, testosterone, and 11-ketotestosterone, but a significant threefold increase in 17-hydroxyprogesterone was detected in PNX-14 treated fish (P = 0.049, Fig. 7).
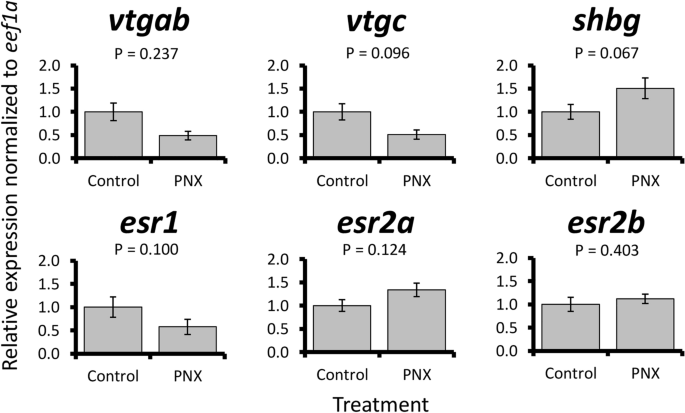
Relative mRNA expression normalized to eef1a of six steroid-associated liver genes (vtgab, vtgc, shbg, esr1, esr2a, and esr2b) in control and PNX-treated puffers. Each bar represents the mean ± standard error, and significance was assessed at P < 0.05.
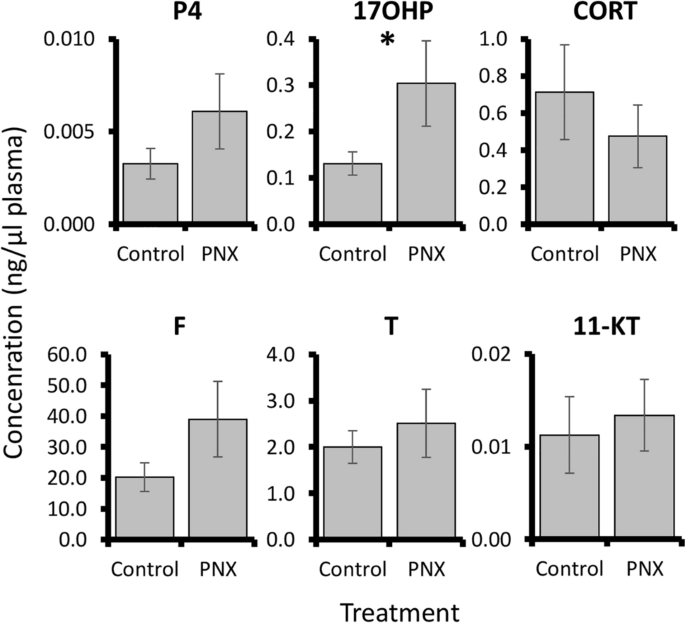
Blood plasma concentrations (ng/μl plasma) of six steroids (P4, progesterone; 17OHP, 17-hydroxyprogesterone; CORT, corticosterone; F, cortisol; T, testosterone; 11-KT, 11-ketotestosterone). Each bar represents the mean ± standard error and the asterisk indicates significance (P = 0.049).
Discussion
PNX-14 exhibited pleiotropic actions in puffer, ranging from immune regulation to effects on cell growth, glucose metabolism, and reproductive processes. In particular, the most dramatic response in both ovary and hypothalamus was related to immunity, although broad changes differed between organs and were largely anti- or pro-inflammatory, respectively. PNX roles as an anti-inflammatory peptide are just beginning to be understood, including suppressive effects on cytokines and inflammation mediators in a variety of cell types19,40,41,42,43. Across these studies, many observed effects were similar to ovary changes in puffer, such as decreases in interleukin-1β, interleukin-6, toll-like receptor 4, and nuclear factor kappa B-associated pathways. The suppressive effects of PNX on inflammation processes may be highly conserved across vertebrates; however, this is also likely context dependent, since these same pathways were also detected within the hypothalamus, but were upregulated. The pro-inflammatory hypothalamic signal may instead be associated with energy metabolism and the gut–brain axis. For instance, hypothalamic inflammation is known to affect energy balance and contribute to insulin resistance44,45. Indeed, administration of another gut-associated peptide (glucose-dependent insulinotropic peptide, GIP) can both increase interleukin-1β and interleukin-6 in the mouse hypothalamus and impair insulin effects45. The putative PNX receptor, SREB3, has also been implicated in inflammatory roles within the mammalian brain, where the receptor is both critical to phoenixin effects in both astrocytes and microglia, and can be induced by an interleukin19,46. Spatially, SREB3 expression has been confirmed within the supraoptic and paraventricular nuclei of the hypothalamus in several mammals47, while PN...
Comments
Post a Comment