Diversity of myxozoans (Cnidaria) infecting Neotropical fishes in ... - Nature.com
Abstract
Myxozoans are a unique group of microscopic parasites that infect mainly fishes. These extremely reduced cnidarians are highly diverse and globally distributed in freshwater and marine habitats. Myxozoan diversity dimension is unknown in Mexico, a territory of an extraordinary biological diversity. This study aimed to explore, for the first time, myxozoan parasite diversity from fishes of the Neotropical region of Mexico. We performed a large morphological and molecular screening using host tissues of 22 ornamental and food fish species captured from different localities of Veracruz, Oaxaca and Chiapas. Myxozoan infections were detected in 90% of the fish species, 65% of them had 1 or 2 and 35% had 3 and up to 8 myxozoan species. Forty-one putative new species were identified using SSU rDNA phylogenetic analyses, belonging to two main lineages: polychaete-infecting (5 species) and oligochaete-infecting (36 species) myxozoans; from those we describe 4 new species: Myxidium zapotecus sp. n., Zschokkella guelaguetza sp. n., Ellipsomyxa papantla sp. n. and Myxobolus zoqueus sp. n. Myxozoan detection increased up to 6 × using molecular screening, which represents 3.7 × more species detected than by microscopy. This study demonstrated that Neotropical fishes from Mexico are hosts of a multitude of myxozoans, representing a source of emerging diseases with large implications for economic and conservation reasons.
Introduction
Myxozoans are microscopic cnidarian parasites that are globally distributed in marine and freshwater habitats. These spore-forming parasites are strongly reduced in size and morphology when compared to their free-living cnidarian relatives. Myxozoans have complex life cycles that involved annelids and bryozoans as definitive invertebrate hosts and predominantly fishes as intermediate vertebrate hosts. In both hosts, waterborne, infectious spores are formed; actinospores in the invertebrate host and myxospores in the vertebrate host. These parasites are particularly well known for the disease they cause in aquaculture and wild fish stocks1.
Myxozoans are a highly diverse group, with > 2400 species, representing approximately one fifth of the phylum Cnidaria2,3. However, the biodiversity of myxozoan species remains unknown and is very likely underestimated4, especially in certain geographic areas. Southern Mexico is included in the so-called Mesoamerican biodiversity hotspot, which includes areas that have high concentrations of endemic species and that are experiencing exceptional loss of habitat5. Myxozoans are considered an understudied group in Mexico, especially when compared to other parasitic groups with long tradition in the Mexican fish parasitology research6,7. So far, only six species of myxozoans have been reported in Mexico, three from marine habitats, i.e. Kudoa dianae Dyková, Fajer Avila & Fiala, 2002, Myxobolus mexicanus Yoshino & Noble, 1973 and Myxidium coryphaenoidium Noble, 1966, and three from freshwater habitats i.e. Myxobolus nuevoleonensis Salinas, Jiménez-Guzmán, Galaviz-Silva & Ramírez-Bon, 1991, Myxobolus cartilaginis Hoffman, Putz & Dunbar, 1965 and Henneguya exilis Kudo, 19298,9,10,11,12,13,14. From these, only K. dianae and M. coryphaenoidium have molecular data available13,15. Additionally, a myxozoan infection episode in tilapia was reported in Mexico, without species identification of the causative agent16.
During wild fish collection campaigns in the neotropical region of Mexico, myxozoan infections were detected. This study seeks to explore myxozoan species diversity from fishes in southern Mexico, using morphological and molecular tools. We provide the morphological and molecular characterization of four new myxozoan species, as well as their phylogenetic relationships. Using host tissues, hidden diversity and occurrence of mixed infections were further investigated through a large-scale molecular screening using PCR techniques and Sanger sequencing. This is the first comprehensive study on the diversity of this group of parasitic cnidarians in Mexico.
Materials and methods
Fishes and samples collection
A total of 120 fish specimens belonging to 22 species, 10 families and 6 orders, were captured using electrofishing and gillnets from 17 localities in 3 states in southern Mexico: Veracruz, Oaxaca and Chiapas (Fig. 1, Supplementary Data 1) between March 2014 and March 2015. In addition, one Atlantic bonito Sarda sarda was purchased in the local market of Alvarado, Veracruz in May 2014. Captured fish were immediately transported to the field laboratory in tanks containing aerated water from the original locality. Fish were euthanized by neural pithing, dissected and species were identified following17,18. Gall bladder was punctured, and bile collected in a tube. Fresh bile smears were examined under the microscope and digital images of the myxospores obtained from fresh material during fieldwork when possible. Aliquots of each bile sample were fixed for molecular (99% ethanol) and morphological analyses (10% neutral buffered formalin). Additionally, pieces of intestine and kidney of nine fish specimens (Intestine and kidney: Mayaheros urophthalmus (n = 2), Parachromis friedrichsthalii (n = 1), Vieja fenestrata (n = 1), Rhamdia guatemalensis (n = 3); only intestine—Poecilia sphenops (n = 1), Dormitator maculatus (n = 1); only kidney—Dajaus monticola (n = 1), Paraneetroplus bulleri (n = 1); Table 1) were examined under the microscope and preserved in absolute ethanol for molecular screening. In order to avoid DNA cross contamination, scissors and tweezers were flame sterilized during sampling. Fish names, habitats and human uses follow FishBase19. The sampling in this work complies with the current laws and animal ethics regulations of Mexico. Fishes were collected under the Cartilla Nacional de Colector Científico (FAUT 0202) issued by the Secretaría del Medio Ambiente y Recursos Naturales (SEMARNAT), to M.G.V.
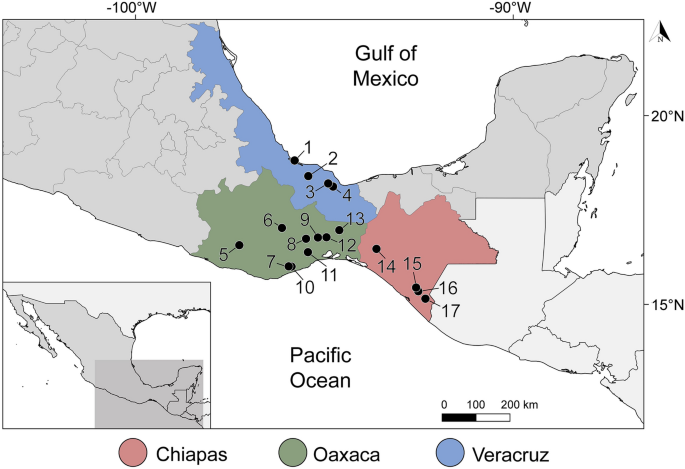
Sampling localities of freshwater and marine fishes along the Neotropical Region of Mexico. Coordinates are provided in Supplementary Data 1. Legend: 1, Alvarado, Veracruz; 2, Tlacotalpan, Veracruz; 3, Catemaco, Veracruz; 4, Río La Palma, Veracruz; 5, El Toronjo, Oaxaca; 6, Río Grande, Mitla, Oaxaca; 7, Río los Sabinos, Oaxaca; 8, Río los Perros, Santa María, Oaxaca; 9, Santa María, Guienagati, Oaxaca; 10, Río Chacalapa, Oaxaca; 11, Río Tequisistlán, Oaxaca; 12, Rio Grande, Matías Romero, Oaxaca; 13, Río Negro, Santa María Chimalapa, Oaxaca; 14, Río San Juan, Cristóbal Colón, Chiapas; 15, Río San Juan, Cristóbal Obregón, Chiapas; 16, Nueva Francia, Chiapas; 17, El Triunfo, Chiapas; 18, Río Huixtla, Chiapas. The map was generated using QGIS v.3.32 (Free and Open Source Geographic Information System, https://www.qgis.org/en/site/#).
Morphological analyses
Digital images of plasmodia and myxospores were taken using a Leica DM750 microscope and Leica ICC50 HD digital camera (1000×) for fresh material during field work; an Olympus BX51 microscope and an Olympus DP70 digital camera (1000×) was used for formalin-fixed material at the lab. Myxospores measurements (n = 162) followed recommendations of20 but using the term polar tubule instead of polar filament (see21). Measurements were taken from digital images using ImageJ 1.47v22 and are given in µm as the mean ± standard deviation with the range in parentheses. Myxospores were air dried directly onto glass slides, stained with Epredia™ Shandon™ Kwik-Diff™ Stains (Thermo Fisher Scientific; Waltham, Massachusetts) and mounted with Neo-mount™ non-aqueous mounting medium (Merck; Darmstadt, Germany). Archival smears are deposited at the Institute of Biology (Universidad Nacional Autónoma de México, UNAM), Colección Nacional de Helmintos CNHE 11950-11953.
Two bile samples were fixed in 2.5% glutaraldehyde in 0.1M PBS and processed for scanning electron microscopy (SEM), by adhering the myxospores to a poly-D-lysine coated coverslip, followed by postfixation and dehydratation according to23. Critical point drying and gold sputtered-coating was performed at the Laboratory of Electron Microscopy, Institute of Parasitology, Czech Academy of Sciences, Czech Republic. Samples were examined using a JEOL JSM-7401 F (JEOL Ltd., Japan) electron microscope.
Molecular analyses
Bile (n = 102), intestine (n = 9) and kidney (n = 9) samples were used for total DNA extraction. Bile content was centrifuged for 5 min at 14.000 RPM to form a pellet and ethanol was removed. Pellet/tissue was airdried until complete ethanol evaporation and resuspended in TNES -Urea (10 mM Tris–HCl (pH 8), 125 mM NaCl, 10 mM EDTA, 0.5% SDS, 4M urea), digested with 100 µg/ml of proteinase K, overnight at 55 °C, and extracted following a standard phenol–chloroform protocol24. The extracted DNA was re-suspended in 30–100 µL RNAse/DNAse-free water. Small subunit ribosomal SSU rDNA amplicons were first amplified using primers ERIB1 (5′-ACC TGG TTG ATC CTG CCA G-3′) and ERIB10 (5′-CTT CCG CAG GTT CAC CTA CGG-3')25 followed by a nested PCR with Myxgp2f (5′-WTG GAT AAC CGT GGG AAA-3′)26 and ACT1r (5′-AAT TTC ACC TCT CGC TGC CA-3′)27. This PCR protocol was used to screen all samples obtained in this study. PCRs were conducted in 10 μl reactions with 0.025 Uμl−1 TITANIUM Taq DNA polymerase and 10 × buffer which contained 1.5 mM MgCl2 (BD Biosciences Clontech), with 0.2 mM of each dNTP, 0.5 µM of each primer, and 10–150 ng of template DNA or 0.5 µL of PCR product. Cycling conditions of initial PCR consisted of 95 °C for 2 min, followed by 35 cycles of 95 °C for 50 s, 60ºC for 50 s and 68 °C for 2 min, and final extension 68 °C for 4 min. Nested PCR consisted of 95 °C for 3 min, followed by 30 cycles of 95 °C for 50 s, 58 °C for 50 s and 68 °C for 1 min 20 s with final extension 68 °C for 4 min. Expected DNA amplicons (≈900–1000 bp) were visualized in 1% agarose gel in sodium acetate buffer and purified for sequencing using a Gel/PCR DNA Fragments Extraction Kit (Geneaid Biotech Ltd., USA). Preferably, direct sequencing of PCR products using nested PCR primers was attempted. Additional primers were used to obtain longer fragments of the SSU rDNA region for some myxospore positive samples (Supplementary Data 2). Problematic amplicons (double peaks, poor quality sequences) were cloned into the pDrive Cloning vector (Qiagen PCR Cloning Kit, Germany) and transformed into the competent E. coli strain XL-1. Plasmid DNA was isolated using a High Pure Plasmid Isolation Kit (Roche Applied Science, Germany) and sequenced using primers M13F (5′-GTA AAA CGA CGG CCA G-3′) and M13R (5´- AAC AGC TAT GAC CAT G -3´). Sequences were obtained with an ABI PRISM 3130 × 1 automatic sequencer (Applied Biosystems; Prague, Czech Republic). The overlapping partial sequences of SSU rDNA were trimmed and assembled into single consensus contigs using Geneious Prime 2022.2 (Biomatters; Auckland, New Zealand; https://www.geneious.com) and submitted to GenBank (accession numbers OQ888222-OQ888300; see Supplementary Data 3).
Phylogenetic analyses
To determine the phylogenetic relationships of identified myxozoans, two sequence alignments were performed, one for myxozoans clustering in the oligochaete-infecting lineage and a second for myxozoans clustering in the polychaete-infecting lineage (see28). Each alignment included the newly generated SSU rDNA sequences as well as sequences of closely related myxozoans available in GenBank, which were retrieved using BLASTN. Nucleotide sequences were aligned using MAFFT version 7.490 (29) implemented in Geneious Prime R11.1 (https://www.geneious.com), using the E-INS-i algorithm, with a gap opening penalty of 2.0. Alignments were edited to remove highly variable sections. Maximum likelihood (ML) analyses were performed using RAxML v7.2.830 with the GTR + Γ model of nucleotide substitution with alpha parameter 0.5895 for the oligochaete-infecting lineage and 0.4924 for the polychaete-infecting lineage. The model was selected using jModelTest v2.1.1031. Bootstrap support values were calculated from 1000 replicas. Bayesian inference (BI) was performed using MrBayes v3.032 with the GTR + Γ model of evolution and the same alpha parameters as for ML. MrBayes was run to estimate posterior probabilities over two million generations via two independent runs of four simultaneous Markov Chain Monte Carlo (MCMC) algorithms with every 100th tree saved. Tracer v1.4.133 was used to set the length of the burn-in period and to identify potential convergence issues. Species-specific divergences were identified from proportional distances (in %) calculated in Geneious Prime based on the dataset used for the ML analysis.
Results
Prevalence and detection
Microscopic examination showed that 16% (17/106) of the gall bladders were positive for myxosporean infection (Table 1). Myxozoan prevalence based on microscopic observations ranged from 14.3 to 100% among fish species. Eleven different species/morphotypes could be detected (Tables 1 and 2, Figs. 2 and 3), four of which are described below. Molecular screening revealed that 49% (50/102) of the gall bladder samples were positive for myxozoans, with prevalence of infection ranging from 25 to 100% among the analyzed fish species. None of the intestine and kidney samples were microscopically positive. Molecularly, 33.3% (3/9) of intestine and 11.1% (1/9) kidney samples were positive.

Myxospores morphology of the newly described myxozoan species of Neotropical fishes from Mexico. (a–h) Myxospores of Myxidium zapotecus sp. n. (a–c) Valvular view, showing myxospore width variability and surface ridges, (d) sutural view, (e–f) surface ridges pattern in valvular (e) and sutural (f) views and (g–h) schematic drawings of the myxospore, including ornamented valve, valvular views; (i–m) Myxospores of Zschokkella guelaguetza sp. n. (i-j) Surface ridges and suture, (k) sutural view, two different planes, (l) sutural (left) and valvular (right) views of myxospores and (m) schematic drawing of the myxospore, sutural view; (n–r) Plasmodia and myxospores of Ellipsomyxa papantla sp. n. (n) Disporic plasmodia, (o) myxospore, sutural view, (p) myxospore, valvular view, opposed extruded polar filaments, (q) myxospore, sutural view, opposed extruded polar filaments and (r) schematic drawing of the myxospore, sutural view; (s–u) Myxospores of Myxobolus zoqueus sp. n. (s) Two myxospores in valvular view, (t) five myxospores in several orientations and (u) schematic drawing of the myxospore, valvular view. a-d, i-l, t: 10% formalin fixed myxospores; n–q, s: fresh myxospores. a–d, i–l, n–q, s–t: light microscopy, scale bar: 5 µm; e–f: fixed in 2.5% glutaraldehyde in 0.1 M PBS, scanning electron microscopy, scale bar: 1 µm; g–h, m, r, u: drawings, scale bar: 2 µm. Suture—head arrow, extruded polar filament—arrow.
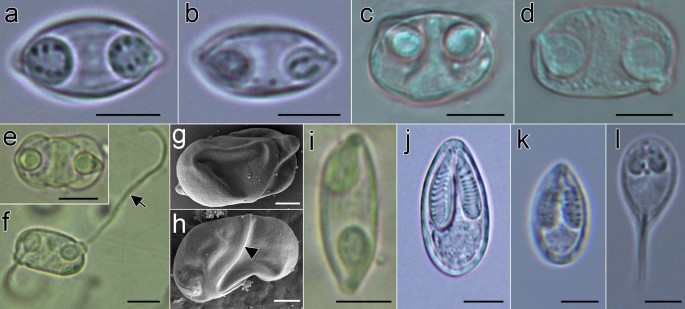
Myxospores morphology of undescribed species and mixed infections of Neotropical fishes from Mexico. (a-b) Mixed infection in Awaous banana (N19), (a) Myxidium zapotecus sp. n., (b) Zschokkella sp. ex A. banana; (c-d) Mixed infection in Dormitator maculatus gall bladder (P5), (c) Zschokkella sp. ex D. maculatus, (d) Ellipsomyxa papantla sp. n.; (e–h) Ellipsomyxa sp. ex Dormitator maculatus (L4), (e) valvular view, (f) sutural view with opposed extruded polar filaments, (g) valvular view (partially collapsed valve), showing valvular protrusions associated to both polar capsules pointing out in opposite directions and (h) sutural view of smooth valves, with small valvular protrusion, where the opening and tip of the polar capsule its located; (i) Myxidium sp. ex Paraneetroplus sp. (C12); (j) Myxobolus sp. ex Cichlasoma urophthalmus (N1); (k) Myxobolus sp. ex Profundulus punctatus (N23); (l) Henneguya sp. ex Parachromis friedrichsthalii (N13). a–b, k–l: fresh myxospores; c–d, e–f, i–j: 10% formalin fixed myxospores; a–f, i–l: light microscopy, scale bar: 5 µm; g–h: fixed in 2.5% glutaraldehyde in 0.1M PBS, Scanning electron microscopy, scale bar: 2 µm. Suture—head arrow, extruded polar filament—arrow.
The most frequently collected fish host species were three cyprinodontiforms, Profundulus punctatus (Profundulidae) (n = 23), Profundulus oaxacae (n = 14) and Poecilia mexicana (Poeciliidae) (n = 8); one mugiliform, Dajaus monticola (Mugilidae) (n = 7); and one perciform, Dormitator maculatus (Eleotridae) (n = 11). Myxozoan prevalence in the gall bladder of these hosts ranged from no detection to 27.3% microscopically, and from 25 to 85.7% by PCR detection (Table 1). Congruent prevalence of infection between microscopic examination and molecular screening was only observed in Awaous banana (Gobiidae) (100%) i.e., all three specimens analyzed showed myxospores in the bile. In some cases, microscopy did not reveal infection by myxospores, but PCR detected the presence of myxozoans up to 66% in some fish host species (e.g., 66.7% in Mayaheros urophthalmus (Cichlidae), 50% in Poecilia mexicana, see Table 1). Overall, molecular screening increased myxozoan detection rates by 1.7 × to 6×. The only two myxozoan negative fish species were Tlaloc labialis (Profundulidae) (n = 1) and Cichlasoma trimaculatum (Cichlidae) (n = 1).
Description of new species
Phylum Cnidaria Hatschek, 1888.
Unranked subphylum Myxozoa Grassé, 1970.
Class Myxosporea Bütschli, 1881.
Order Bivalvulida Schulman, 1959.
Suborder Variisporina Lom et Noble, 1984.
Family Myxidiidae Thélohan, 1892.
Genus Myxidium Bütschli, 1882.
Myxidium zapotecus sp. n
Description of myxospores
Based on 18 myxospores fixed in formalin from the bile of one host (code: C8). Data obtained using light microscopy and SEM. Myxospores fusiform in valvular view, ellipsoidal in sutural view (Fig. 2a–h) measuring 12.5 ± 1.3 (10.8–14.6; n = 18) in length, 8.6 ± 1.2 (6.9–11.3; n = 10) in width and 6.3 ± 0.5 (5.6–7.1; n = 8) in thickness, with pointed ends. Two valves with 9–10 longitudinal surface ridges (Fig. 2e, f and h), parallel to sutural plane. Suture bisects the myxospore. Two equal subspherical polar capsules, 4.2 ± 0.6 (3.1–5.4; n = 35) long and 3.4 ± 0.6 (2.6–4.6; n = 35) wide (Fig. 2a–d, g). Polar capsules positioned at opposite ends of the myxospore and opening to different sides in sutural plane. Polar tubule arranged in 3 coils. Sporoplasm binucleate, in middle of myxospore.
Taxonomic summary
Type host: River goby Awaous banana (Valenciennes) (Gobiiformes: Gobiidae).
Type locality: Río Negro, Santa María Chimalapa (16°53'55''N; 94°41'37'' W), Oaxaca, Mexico.
Other locality: Río Grande, Matías Romero (16°47'29"N; 95°01'02"W), Oaxaca, Mexico.
Additional hosts and localities: Mountain mullet Dajaus monticola (Bancroft) (Mugiliformes: Mugilidae) ex Río Grande, Matías Romero (16°47'29"N; 95°01'02" W), Oaxaca, Mexico; Astyanax sp. (Characiformes: Characidae) ex Río Negro, Santa María Chimalapa (16°53'55'' N; 94°41'37'' W), Oaxaca, Mexico.
Site in hosts: Gall bladder.
Prevalence in type host: 66.6 % (2/3) microscopic detection (1/1—Type locality, 1/2—Other locality), 66.6% (2/3) molecular detection.
Prevalence in other hosts: 33.3% (1/3) in Dajaus monticola and 33.3% (1/3) in Astyanax sp. (molecular detection only).
Material deposited: Kwik-Diff stained slides of air-dried myxospores (C8) (CNHE 11950).
Representative SSU rDNA sequences: GenBank OQ888226 ex Awaous banana (1,806 bp; code: C8); OQ888240 ex A. banana (919 bp; code HP179_N19); OQ888239 ex Dajaus monticola (920 bp; code: HP183_N24); OQ888242 and OQ888241 ex Astyanax sp. (919 and 919 bp; code: HP121_C10 & HP123_C10).
ZooBank registration: The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub:D74C1323-E34D-4D7A-816E-098EA48188C4. The LSID for the new name Myxidium zapotecus sp. n. is urn:lsid:zoobank.org:act: 46A34393-6842-4319-87EF-593D17A69837.
Etymology: The species is named after the indigenous pre-Columbian civilization in the Valley of Oaxaca in Mesoamerica, the Zapotecs.
Remarks
Myxospore measurements of Myxidium zapotecus sp. n. from another specimen of Awaous banana (N19) are provided in Table 2 (Fig. 3a). In this host, a mixed infection was detected with a Zschokkella sp. (Fig. 3b). Myxidium zapotecus sp. n. has overall myxospore morphology characteristic of the genus Myxidium. For differential diagnosis we selected species of Myxidium reported from North America, from other gobiids and other species that showed sequence similarity to our new species34 (Supplementary Data 4). The new species can be distinguished from its congeners based on the fish intermediate host and/or geographic location. Myxidium zapotecus sp. n. differs in myxospore shape to all the species compared, except for Myxidium phyllium (Davis, 1917), Myxidium truttae Léger, 1930 and Myxidium eminentis Ishizaki, 1957. These four species shared a fusiform shape in valvular view, in contrast to the other species with oval, rounded, more ellipsoid myxospores. The new species differs in both myxospore length and width to Myxidium coryphaenoidium Noble, 1966, Myxidium amazonense Mathews, Silva, Maia & Adriano, 2015 and Myxidium whippsi McAllister, Cloutman, Leis & Robison, 2022, and in myxospore width to Myxidium pseudocuneiforme Chen, Zhang, Whipps, Yang & Zhao, 2021 and Myxidium kudoi Meglitsch, 1937. It differs in polar capsule shape to Myxidium glossogobi Chakravarty, 1939 and Myxidium pseudocuneiforme (subspherical vs pyriform). The new species differs in number of polar tubule coils to Myxidium coryphaenoidium, Myxidium amazonense and Myxidium pseudocuneiforme (3 coils vs. > 4 coils; Supplementary Data 4) and in number of valve striations to Myxidium pseudocuneiforme (9–10 vs. 6–8), Myxidium whippsi (9–10 vs. 5) and Myxidium glossogobi (9–10 vs. no striations).
The new species overlaps in myxospore size with Myxidium phyllium, Myxidium truttae and Myxidium eminentis. However, Myxidium zapotecus sp. n. has longer and wider myxospores and longer polar capsules than Myxidium phyllium and Myxidium truttae and differs in fish host (river goby Awaous banana vs mosquitofish Gambusia affinis and brown trout Salmo trutta, respectively) and in geographic location (Mexico vs USA and France, respectively). Myxidium zapotecus sp. n. differs from Myxidium eminentis in myxospore width, fish host (river goby Awaous banana vs flat-headed goby Luciogobius guttatus) and geographic location (Mexico vs Japan).
Genus Zschokkella Auerbach, 1910.
Zschokkella guelaguetza sp. n
Description of myxospores
Based on 25 formalin fixed myxospores from one host (code: N26) by light microscopy. Myxospores ellipsoidal in sutural view and slightly semicircular in valvular view (Fig. 2i–m) measuring 10.9 ± 0.4 (9.1–11.9; n = 25) in length, 6.2 ± 0.6 (5.6–7.5: n = 16) in width, 7.0 ± 0.4 (6.1–7.3, n = 9) thickness, with blunted ends. Two unequal valves, with longitudinal surface ridges, sinuous suture (Fig. 2i–k). Two equal subspherical polar capsules, 3.2 ± 0.3 (2.6–3.7: n = 50) long and 2.8 ± 0.2 (2.2–3.2: n = 50) wide. Polar capsules subterminal to one side/valve (Fig. 2m). Polar tubule arranged in 2–3 coils. Sporoplasm binucleate, in middle of myxospore.
Taxonomic summary
Type host: River goby Awaous banana (Valenciennes) (Gobiiformes: Gobiidae).
Type locality: Río Grande, Matías Romero (16°47'29"N; 95°01'02"W), Oaxaca, Mexico.
Other host and locality: Astyanax sp. (Characiformes: Characidae) ex Río Negro, Santa María Chimalapa (16°53'55''N; 94°41'37''W), Oaxaca, Mexico.
Site in hosts: Gall bladder.
Prevalence in type host: 33.3% (1/3) microscopic detection, 33.3% (1/3) molecular detection.
Prevalence in other host: 33.3% (1/3) in Astyanax sp. (molecular detection only).
Material deposited: Kwik-Diff stained slides of air-dried myxospores (N26) (CNHE 11951).
Representative SSU rDNA sequences: GenBank OQ888223 ex Awaous banana (1,926 bp; code: N26); OQ888237 ex Astyanax sp. (993 bp; code: HP122_C10).
ZooBank registration: The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub: D74C1323-E34D-4D7A-816E-098EA48188C4. The LSID for the new name Zschokkella guelaguetza sp. n. is urn:lsid:zoobank.org:act: 2BC5693F-D700-4B00-8D76-46E0C467B55E.
Etymology: The species is named after the colorful festivity of La Guelaguetza, an annual indigenous cultural dancing event that takes place in Oaxaca, Mexico.
Remarks
The novel species has overall myxospore morphology characteristics of the genus Zschokkella. For differential diagnosis we selected species of Zschokkella reported in gobiids and other fish species that showed sequence similarity to the new species35. Zschokkella guelaguetza sp. n. can be distinguished from other Zschokkella species by fish alternate host and geographical location (Supplementary Data 4). The new species differs in myxospore shape to Zschokkella fujitai Ozaki & Isizaki, 1941, Zschokkella glossogobii Kalavati & Vaidehi, 1991, Zschokkella nova Klokačewa, 1914 and Zschokkella trachini Azizi, Rangel, Castro, Santos & Bahri, 2016 (Ellipsoidal vs Ovoid, oval or elongate myxospore in sutural view). It differs in myxospore length and width to Zschokkella trachini and Zschokkella soleae Yemmen, Marton, Bahri & Eszterbauer, 2013, and in myxospore length to Zschokkella fujitai. The new species differs in polar capsule shape to all species (i.e. subspherical vs pyriform, ovoid or spherical), except to Zschokkella auratis Rocha, Casal, Rangel, Severino, Castro, Azevedo & Santos, 2013, Zschokkella fujitai and Zschokkella balistoidi Heiniger & Adlard, 2014. Zschokkella guelaguetza sp. n. differs in polar capsule length and width to Zschokkella fujitai and to Zschokkella glossogobii and in number of polar tubule coils to Zschokkella soleae and Zschokkella auratis (2 to 3 coils vs 4 to 5 coils).
Zschokkella guelaguetza sp. n. overlaps in myxospore size with Zschokkella balistoidi and Zschokkella gobidiensis Sarkar & Ghosh, 1991. The new species has slightly longer and wider polar capsules than Zschokkella balistoidi and differs in host (river goby Awaous banana vs titan triggerfish Balistoides viridescens) and geographic location (Mexico vs Australia). Zschokkella guelaguetza sp. n. can be differentiated from Zschokkella gobidiensis on having shorter and narrower myxospores, on fish intermediate host (river goby Awaous banana vs tank goby Glossogobius giuris) and on geographic location (Mexico vs India).
Family Ceratomyxidae Doflein, 1899.
Genus Ellipsomyxa (Køie, 2003).
Ellipsomyxa papantla sp. n
Description of myxospores
Based on 19 fresh myxospores from one host (code: N4) by light microscopy. Myxospores ovoid in valvular view and ellipsoidal in sutural view (Fig. 2n–r) measuring 12.9 ± 0.8 (11.6–15.0; n = 19) in length, 9.1 ± 0.5 (7.6–9.9; n = 7) in width and 7.3 ± 0.7 (6.1–8.2; n = 12) in thickness. Two smooth valves, transverse suture, forming acute angle to thickness (Fig. 2o). Small valvular protrusions associated with tips of polar capsules (Fig. 2p,q). Two equal spherical to pyriform polar capsules, 3.8 ± 0.5 (2.6–4.6; n = 25) long and 3.3 ± 0.5 (2.2–4.2: n = 25) wide. Polar capsules close to sutural plane, discharging at opposite sides (Fig. 2p,q). Polar tubule measuring 20.9–29.2 in length (n = 6), arranged in 3–4 coils. Sporoplasm binucleate, in middle of myxospore. Disporic plasmodia (Fig. 2n).
Taxonomic summary
Type host: Fat sleeper Dormitator maculatus (Bloch) (Gobiiformes: Eleotridae).
Type locality: Tlacotalpan (18º36'41''N; 95º39'44''W), Veracruz, Mexico.
Site in host: Gall bladder.
Prevalence: 18.2% (2/11) microscopic detection, 30% (3/10) molecular detection.
Material deposited: Kwik-Diff stained slides of air-dried myxospores (N4) (CNHE 11952).
Representative SSU rDNA sequences: GenBank OQ888230 ex Dormitator maculatus (1,603 bp; code: N4); OQ888285 ex D. maculatus (761 bp; code: HP92_J5), and OQ888286 ex D. maculatus (761 bp; code: HP89_P5).
ZooBank registration: The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub: D74C1323-E34D-4D7A-816E-098EA48188C4. The LSID for the new name Ellipsomyxa papantla sp. n. is urn:lsid:zoobank.org:act: F901367A-FCBE-44F1-8392-026367CEA598.
Etymology: The species is named after one of the localities where the ancient Mesoamerican ritual ceremony, "Danza de los Voladores" takes place - Papantla, Veracruz, Mexico.
Remarks
Measurements for formalin-fixed Ellipsomyxa papantla sp. n. myxospores detected in another specimen of D. maculatus (P5) are provided in Table 2 (Fig. 3d). In this host, a mixed infection with a Zschokkella sp. was observed (see Fig. 3c). Ellipsomyxa papantla sp. n. has overall myxospore morphology characteristic of the genus Ellipsomyxa. For differential diagnosis we selected all species of Ellipsomyxa reported to date (Supplementary Data 4). The novel species is unique with respect to all other Ellipsomyxa species in fish host and geographic location. It differs in myxospore shape to all the species compared, except to Ellipsomyxa apogoni Heiniger & Adlard, 2014; both species having ellipsoidal myxospores in sutural view and ovoid in valvular view. Ellipsomyxa papantla sp. n. myxospores differ in length and width to Ellipsomyxa gobioides Azevedo, Videira, Casal, Matos, Oliveira, Al-Quraishy & Matos, 2013 and Ellipsomyxa boleophthalmi Vandana, Poojary, Tripathi, Pavan-Kumar, Pratapa, Sanil & Rajendran, 2021. It differs in myxospore length to Ellipsomyxa fusiformis (Davis, 1917), Ellipsomyxa gobii Køie, 2003, Ellipsomyxa syngnathi Køie & Karlsbakk, 2009, Ellipsomyxa apogoni Heiniger & Adlard, 2014 and Ellipsomyxa tucujuensis Ferreira, da Silva, de Carvalho, Bittencourt, Hamoy, Matos, & Videira, 2021. Ellipsomyxa papantla sp. n. differs in myxospore width to Ellipsomyxa arariensis Da Silva, Matos, Lima, Furtado, Hamoy & Matos, 2018, Ellipsomyxa arothroni Heiniger & Adlard, 2014, Ellipsomyxa kalthoumi Thabet, Tlig-Zouari, Al Omar & Mansour, 2016, Ellipsomyxa manilensis Heiniger & Adlard, 2014 and Ellipsomyxa plagioscioni Zatti, Maia & Adriano, 2020. The new species differs in polar capsule length to Ellipsomyxa arothroni and Ellipsomyxa kalthoumi, and in polar capsule length and width to Ellipsomyxa manilensis. It differs in the number of polar tubule coils to Ellipsomyxa adlardi Whipps & Font, 2013, Ellipsomyxa arariensis, Ellipsomyxa gobii, Ellipsomyxa gobioides, Ellipsomyxa kalthoumi, Ellipsomyxa plagioscioni, Ellipsomyxa mugilis (Sitja-Bobadilla & Alvarez-Pellitero, 1993), Ellipsomyxa nigropunctatis Heiniger & Adlard, 2014, Ellipsomyxa syngnathi and Ellipsomyxa tucujuensis (3 to 4 vs. > 5 coils).
The new species overlaps in myxospore size with Ellipsomyxa amazonensis Zatti, Atkinson, Maia, Correa, Bartholomew & Adriano, 2018, Ellipsomyxa ariusi Chandran, Zacharia, Sathianandan & Sanil, 2020 and Ellipsomyxa paraensis Zatti, Maia & Adriano, 2020. However, Ellipsomyxa papantla sp. n. has longer and wider myxospores than these three species, as well as wider polar capsules, and differs in host range (Fat sleeper Dormitator maculatus vs gilded catfish Brachyplatystoma rousseauxii, threadfin sea catfish Arius arius and tucunaré Cichla monoculus) and geographic location (Mexico vs Brazil and India).
Suborder Platysporina Kudo, 1919.
Family Myxobolidae Thélohan, 1892.
Genus Myxobolus Bütschli, 1881.
Myxobolus zoqueus sp. n
Description of myxospores
Based on 30 fresh myxospores from one host (code: N21) by light microscopy. Myxospores oval in valvular view, ellipsoidal in sutural view (Fig. 2s–u) measuring 7.9 ± 0.3 (7.1–8.6; n = 30) in length, 5.9 ± 0.4 (4.7–6.5; n = 30) in width. Two smooth valves, suture straight. Four to five notches at sutural edge occasionally evident, at posterior part (Fig. 2s,u). Two equal pyriform polar capsules 2.6 ± 0.3 (2.0–3.1: n = 58) long and 1.6 ± 0.2 (1.1–2.2; n = 58) wide, opening at anterior part. Polar tubule arranged in 2–3 coils. Binucleate sporoplasm at posterior part of the myxospore with iodinophilous vacuole.
Taxonomic summary
Type host: Mountain mullet Dajaus monticola (Bancroft) (Mugiliformes: Mugilidae).
Type locality: Río Grande, Matías Romero (16°47'29" N; 95°01'02" W), Oaxaca, Mexico.
Site in host: Gall bladder.
Prevalence: 33.3% (1/3) microscopic detection, 66.6% (2/3) molecular detection.
Material deposited: Kwik-Diff stained slides of air-dried myxospores (N21) (CNHE 11953).
Representative SSU rDNA sequences: GenBank OQ888227 ex Dajaus monticola (1,804 bp; code: N21); and OQ888253 ex D. monticola (863 bp; code: HP182_N24).
ZooBank registration: The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub: D74C1323-E34D-4D7A-816E-098EA48188C4. The LSID for the new name Myxobolus zoqueus sp. n. is urn:lsid:zoobank.org:act: A99526D1-9F6D-421F-A4D9-52C0DEAA60C3.
Etymology: The species is named after the Zoque people, an indigenous ethnic group present in Oaxaca, Chiapas and Tabasco in Mexico.
Remarks
Myxobolus zoqueus sp. n. has overall myxospore morphology characteristics of the genus Myxobolus. For differential diagnosis we selected species of Myxobolus reported from Mexico and from other fish intermediate hosts belonging to the order Mugiliformes (Supplementary Data 4). Myxobolus zoqueus sp. n. can be distinguished from its congeners based on fish host species and site of infection (gall bladder). The new species differs in myxospore length to Myxobolus curema Vieira, Agostinho, Negrelli, da Silva, de Azevedo & Abdallah, 2022, Myxobolus pupkoi Gupta, Haddas-Sasson, Gayer & Huchon, 2022 and Myxobolus nuevoleonensis Segovia-Salinas, Jiménez-Guzmán, Galaviz-Silva & Ramírez-Bon, 1991. It differs in myxospore width to Myxobolus exiguus (Thélohan, 1895), Myxobolus peritonaeum Rocha, Casal, Alves, Antunes, Rodrigues & Azevedo, 2019 and Myxobolus ramadus Rocha, Casal, Alves, Antunes, Rodrigues & Azevedo, 2019. Myxobolus zoqueus sp. n. differs in both polar capsule length and width to Myxobolus cartilaginis Hoffman, Putz & Dunbar, 1965, Myxobolus exiguus, Myxobolus nuevoleonensis and Myxobolus ramadus, and differs in polar capsule length to Myxobolus muscularis Rocha, Casal, Alves, Antunes, Rodrigues & Azevedo, 2019 and Myxobolus peritonaeum Rocha, Casal, Alves, Antunes, Rodrigues & Azevedo, 2019. The new species differs in the number of coils of the polar tubule (2 to 3 vs. > 4 and up to 11) and in the number of sutural folds (4 to 5 vs. > 6 up to 10) to all compared species.
Myxobolus zoqueus sp. n. overlaps in myxospore size to Myxobolus mexicanus Yoshino & Noble, 1973. However, the new species has shorter and narrower myxospores, as well as shorter polar capsules. Myxobolus zoqueus sp. n. also differs in polar capsule positions, which was described as 'anteriorly located and shifted to one side of the myxospore's valvular midline' for Myxobolus mexicanus36. In addition, the new species differ from Myxobolus mexicanus by its fish host (mountain mullet Dajaus monticola vs shoulderspot grenadier Coelorinchus scaphopsis), site of infection (gall bladder vs kidney), geographic location (Oaxaca, southwestern Mexico vs Baja California, northwestern Mexico) and habitat (freshwater vs marine).
Myxobolus zoqueus sp. n. overlaps in myxospore size to the measurements provided for the species Myxobolus cartilaginis ex Micropterus salmoides from Mexico10. The new species is shorter and narrower than Myxobolus cartilaginis ex Micropterus salmoides and the polar capsule length differs between both species. Both species can also be distinguished by its fish host (mountain mullet Dajaus monticola vs largemouth black bass Micropterus salmoides), site of infection (gall bladder vs branchiostegal rays) and geographic location in Mexico (Oaxaca, southwestern Mexico vs Nuevo León, northeastern Mexico).
Molecular and phylogenetic results
A total of 79 novel partial SSU rDNA sequences were generated in this study (Supplementary Data 3). Sixty-seven sequences were representatives of the oligochaete-infecting lineage (mostly freshwater fish infecting species), belonging to three clades: urinary tract clade (11 sequences), biliary tract clade (19 sequences) and Myxobolus clade (37 sequences). Sequences falling in the Myxobolus clade were found to group in subclade I (6 sequences) and VII (31 sequences) (Fig. 4) (subclades according to37). Twelve sequences belonged to the polychaete-infecting lineage (mostly marine fish infecting species), specifically to the biliary tract clade and most likely represented species of Ellipsomyxa Køie, 2003 (Fig. 5). None of the newly generated sequences were conspecific to any species or sequence data available in GenBank.
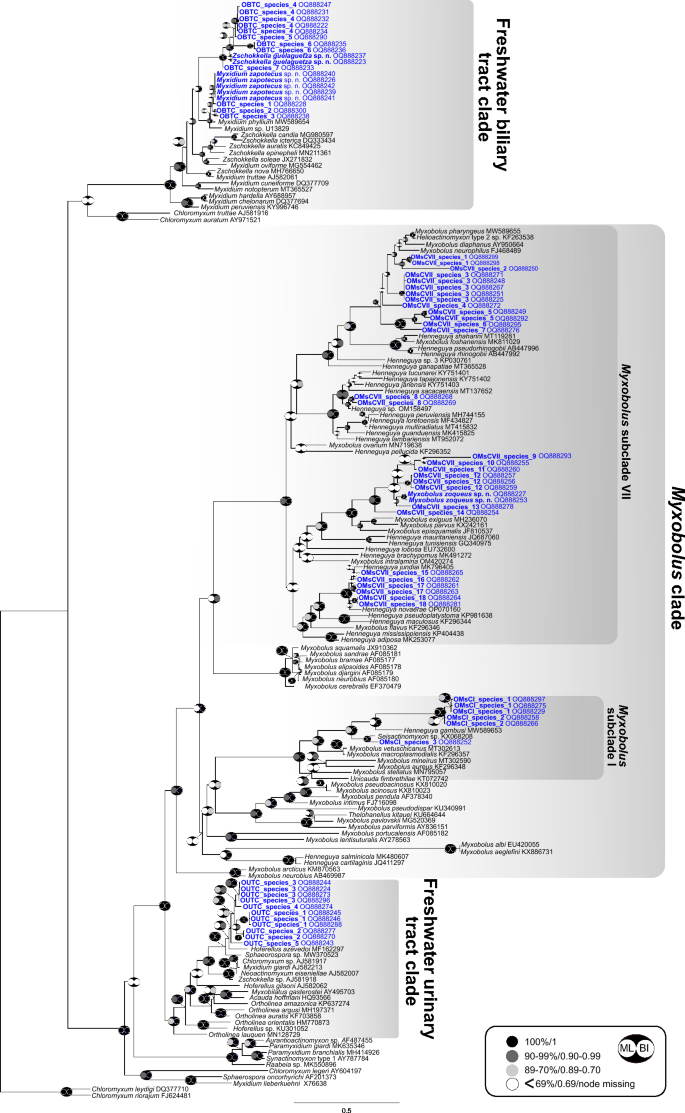
Phylogenetic position of newly identified SSU rDNA myxozoan sequences of Neotropical fishes from Mexico within the oligochaete-infecting lineage. The tree was inferred using maximum likelihood analyses. Bootstrap support and Bayesian posterior probabilities are indicated by pictograms at nodes (see legend). GenBank accession numbers are provided beside taxon names. Newly sequenced taxa are indicated in blue. A scale bar is included to visualize the branch lengths and evolutionary distances between different taxa.